Query Zoo: DSL Gallery for Multilayer Analysis
The Query Zoo is a curated gallery of DSL queries that demonstrate the expressiveness and power of py3plex for multilayer network analysis.
Each example:
Solves a real multilayer analysis problem
Uses the DSL end-to-end with idiomatic patterns
Produces concrete, reproducible outputs
Is fully tested and documented
Why a Query Zoo?
The DSL is most powerful when you see it in action on realistic problems. This gallery shows you how to think about multilayer queries, not just what the syntax is. Use these examples as recipes and starting points for your own analyses.
Overview
The Query Zoo is organized around common multilayer analysis tasks:
Basic Multilayer Exploration — Understand layer statistics and structure
Cross-Layer Hubs — Find nodes that are important across multiple layers
Layer Similarity — Measure structural alignment between layers
Community Structure — Detect and analyze multilayer communities
Multiplex PageRank — Compute multilayer-aware centrality
Robustness Analysis — Assess network resilience to layer failures
Advanced Centrality Comparison — Identify versatile vs specialized hubs
Edge Grouping and Coverage — Analyze edges across layer pairs with top-k and coverage
Layer Algebra Filtering — Use layer set algebra for flexible layer selection
Cross-Layer Paths with Algebra — Find paths while excluding certain layers
Null Model Comparison — Statistical significance testing against null models
Bootstrap Confidence Intervals — Estimate uncertainty in centrality measures
Uncertainty-Aware Ranking — Rank nodes considering variability across layers
All examples use small, reproducible multilayer networks from the examples/dsl_query_zoo/datasets.py module with fixed seeds so you can match the outputs shown here.
Tip
Running the Examples
All query functions are available in examples/dsl_query_zoo/queries.py.
To run all queries and generate outputs:
python examples/dsl_query_zoo/run_all.py
Test the queries with:
pytest tests/test_dsl_query_zoo.py
1. Basic Multilayer Exploration
Problem: You’ve loaded a multilayer network and want to quickly understand its structure. Which layers are densest? How many nodes and edges does each layer have?
Solution: Compute basic statistics per layer using the DSL.
Query Code
Why It’s Interesting
First step in any analysis — Before diving into complex queries, understand your data
Reveals layer diversity — Different layers often have vastly different structures
Identifies sparse vs dense layers — Helps decide which layers need special handling
Example Output
Running on the social_work_network (12 people across social/work/family layers):
Layer |
Nodes |
Edges |
Avg Degree |
|---|---|---|---|
social |
12 |
11 |
1.83 |
work |
11 |
9 |
1.64 |
family |
11 |
6 |
1.09 |
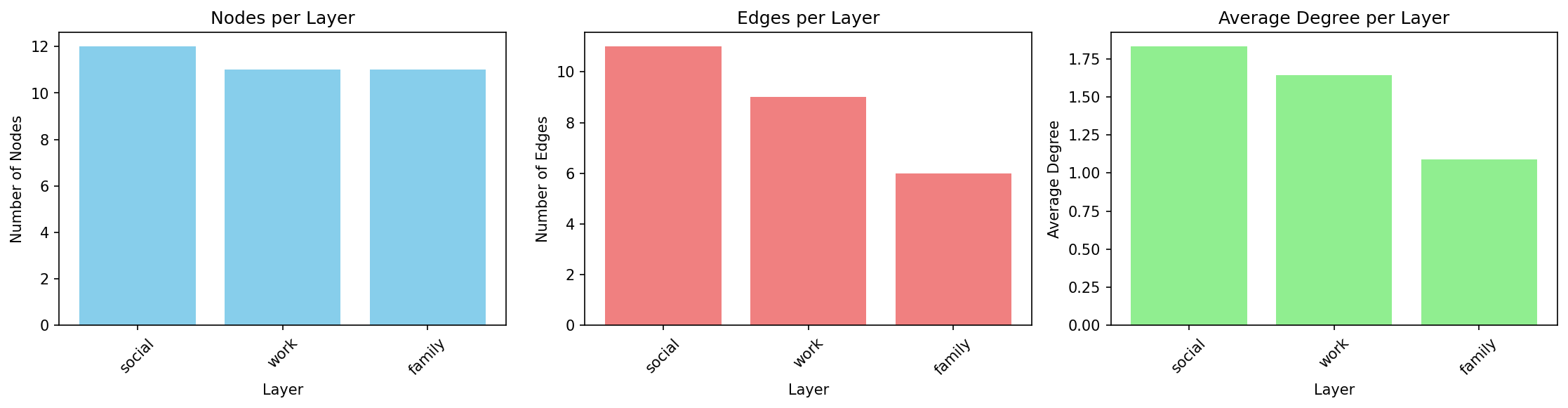
Interpretation: The social layer is densest (highest average degree), while family is sparsest. All layers have similar numbers of nodes, indicating good cross-layer coverage.
DSL Concepts Demonstrated
Q.nodes().from_layers(L[name])— Select nodes from a specific layer.compute("degree")— Add computed attributes to results.execute(network)— Run the query and get results.to_pandas()— Convert to DataFrame for analysis
2. Cross-Layer Hubs
Problem: Which nodes are consistently important across multiple layers? These “super hubs” are critical because they bridge different contexts.
Solution: Find top-k central nodes per layer, then identify which nodes appear in multiple layers’ top lists.
Query Code
Why It’s Interesting
Reveals cross-context influence — Nodes central in one layer might be peripheral in another
Identifies key connectors — Nodes that appear in multiple layers’ top-k are especially important
Robust hub detection — More reliable than single-layer centrality
Example Output
Top cross-layer hubs (k=5):
Node |
Layer |
Degree |
Betweenness |
Layer Count |
|---|---|---|---|---|
Bob |
social |
3 |
0.0273 |
3 |
Bob |
work |
2 |
0.0 |
3 |
Bob |
family |
1 |
0.0 |
3 |
Alice |
work |
3 |
0.0889 |
2 |
Charlie |
social |
3 |
0.0273 |
2 |
Interpretation: Bob appears as a top-5 hub in all three layers (layer_count=3), making him the most versatile connector. Alice and Charlie are hubs in two layers each.
layer_count is the number of distinct layers in which a node enters the per-layer top-k list.
DSL Concepts Demonstrated
.compute("betweenness_centrality", "degree")— Compute multiple metrics at once.order_by("-betweenness_centrality")— Sort descending (-prefix).limit(k)— Take top-k resultsPer-layer iteration and aggregation across layers
3. Layer Similarity
Problem: How similar are different layers structurally? Do they serve redundant or complementary roles?
Solution: Compute degree distributions per layer and measure pairwise correlations.
Query Code
Why It’s Interesting
Detects redundancy — High correlation suggests layers capture similar structure
Guides simplification — Nearly identical layers might be merged
Reveals specialization — Low/negative correlation shows layers serve different roles
Example Output
Correlation matrix for social_work_network:
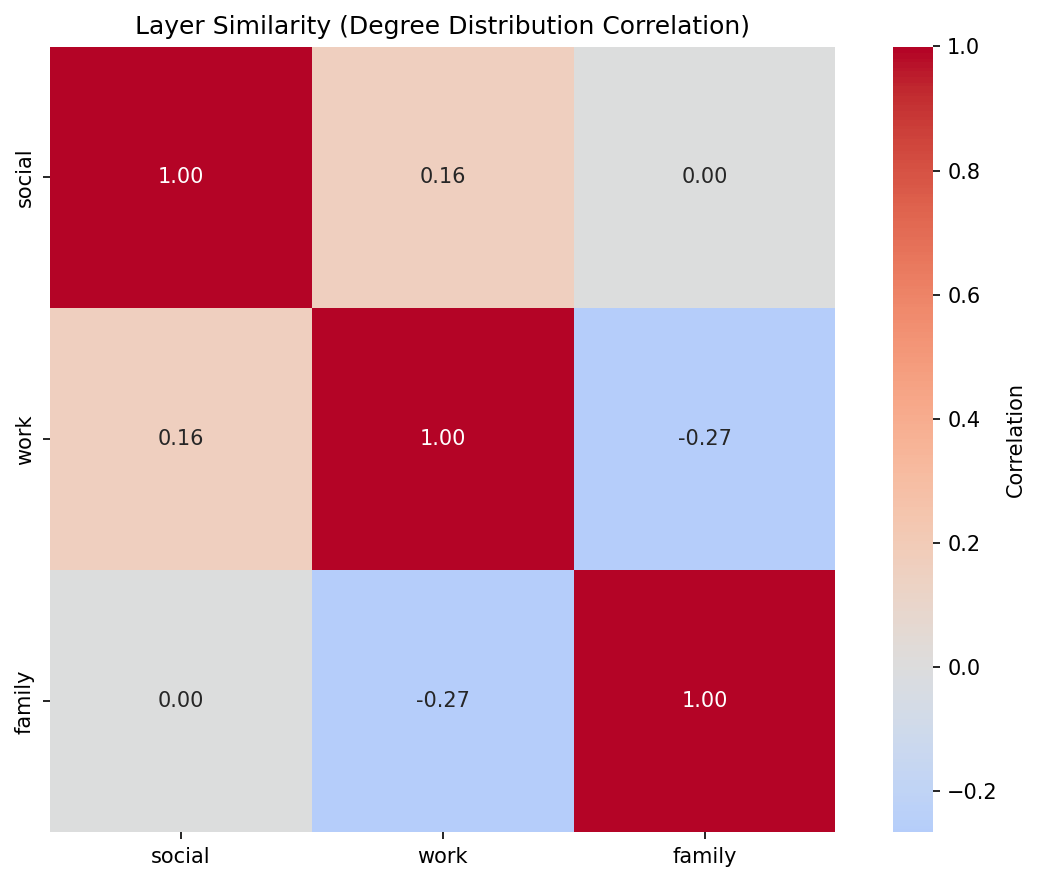
social |
work |
family |
|
|---|---|---|---|
social |
1.000 |
0.159 |
0.000 |
work |
0.159 |
1.000 |
-0.267 |
family |
0.000 |
-0.267 |
1.000 |
Interpretation: Social and work layers have weak positive correlation (0.159), suggesting some structural overlap. Family and work are negatively correlated (-0.267), indicating they capture different connectivity patterns. Correlations are Pearson coefficients computed from the node-by-layer degree matrix. Nodes missing from a layer contribute a degree of 0 in that matrix so every layer uses the same node ordering.
DSL Concepts Demonstrated
Layer-by-layer degree computation
Aggregating results across layers for meta-analysis
Using computed attributes for layer-level comparisons
4. Community Structure
Problem: What communities exist in the multilayer network? How do they manifest across layers?
Solution: Detect communities using multilayer community detection, then analyze their distribution across layers.
Query Code
Why It’s Interesting
Mesoscale structure — Communities reveal organizational patterns
Cross-layer community tracking — See if communities are layer-specific or global
Dominant layers — Identify which layer best represents each community
Example Output
Running on communication_network (email/chat/phone layers):
Community |
Layer |
Size |
Avg Degree |
Dominant Layer |
|---|---|---|---|---|
0 |
10 |
1.8 |
||
1 |
chat |
6 |
2.17 |
chat |
2 |
chat |
3 |
1.67 |
chat |
3 |
phone |
7 |
1.71 |
phone |
Interpretation: Community 0 is email-dominated (10 nodes), while communities 1 and 2 are chat-specific. Community 3 appears primarily in phone communication.
DSL Concepts Demonstrated
Q.nodes().from_layers(L["*"])— Select from all layers.compute("communities")— Built-in community detectionGrouping by
(community_id, layer)for analysisIdentifying dominant layers via aggregation
5. Multiplex PageRank
Problem: Standard PageRank treats each layer independently. How do we compute importance considering the full multiplex structure?
Solution: Compute PageRank per layer, then take the average across layers as a simplified multiplex score. (True multiplex PageRank uses supra-adjacency matrices.)
Query Code
Why It’s Interesting
Multilayer-aware centrality — Accounts for importance across all layers
More robust than single-layer — Averages out layer-specific biases
Essential for multiplex influence — Key for viral marketing, information diffusion
Example Output
Top nodes by multiplex PageRank in transport_network:
Node |
Multiplex PR |
Total Degree |
Bus PR |
Metro PR |
Walking PR |
|---|---|---|---|---|---|
ShoppingMall |
0.1811 |
6 |
0.1362 |
0.1909 |
0.2164 |
Park |
0.1806 |
4 |
0.1449 |
0.0 |
0.2164 |
CentralStation |
0.1683 |
6 |
0.1971 |
0.1909 |
0.117 |
BusinessDistrict |
0.1484 |
4 |
0.079 |
0.1994 |
0.1667 |
Interpretation: ShoppingMall has highest multiplex PageRank (0.1811) because it’s central across all three transport modes. Park has high walking PageRank but zero metro, reflecting its limited accessibility. Scores are the mean of per-layer PageRank values.
DSL Concepts Demonstrated
.compute("pagerank")— Built-in PageRank computationPer-layer iteration with result aggregation
Pivot tables for layer-wise breakdowns
Combining degree and PageRank for richer analysis
6. Robustness Analysis
Problem: How robust is the network to layer failures? What happens if one layer goes offline?
Solution: Simulate removing each layer and measure connectivity loss.
Query Code
Why It’s Interesting
Critical infrastructure identification — Reveals which layers are essential
Redundancy assessment — High robustness indicates good backup coverage
Failure planning — Informs which layers need extra protection
Example Output
Robustness of transport_network:
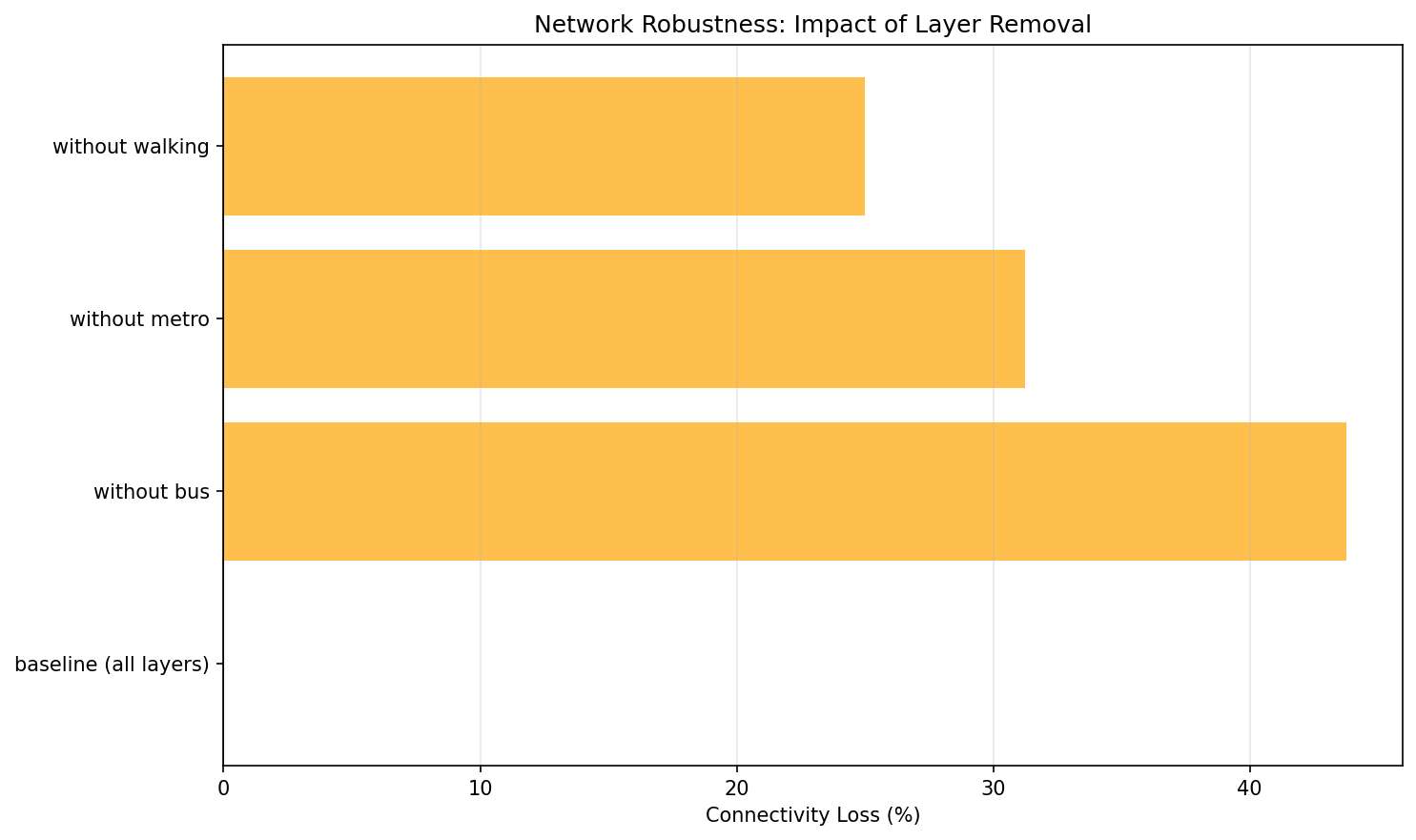
Scenario |
Nodes |
Avg Degree |
Total Edges |
Connectivity Loss (%) |
|---|---|---|---|---|
baseline (all layers) |
14 |
2.14 |
15 |
0.0 |
without bus |
11 |
1.45 |
8 |
46.67 |
without metro |
11 |
1.82 |
10 |
33.33 |
without walking |
14 |
2.0 |
14 |
6.67 |
Interpretation: Removing the bus layer causes 46.67% connectivity loss — it’s the most critical layer. Walking is least critical (only 6.67% loss), indicating good redundancy from other transport modes. Connectivity loss is computed from total degree (divided by 2 for undirected edges), so it assumes undirected layers. The reported loss compares baseline total degree to the degree after removing a layer; for undirected networks total degree is twice the number of edges.
DSL Concepts Demonstrated
Layer algebra:
L["layer1"] + L["layer2"]— Combine layersQ.nodes().from_layers(layer_expr)— Query with dynamic layer selectionsBaseline vs scenario comparison
Measuring connectivity metrics before/after perturbations
7. Advanced Centrality Comparison
Problem: Different centralities capture different notions of importance. Which nodes are “versatile hubs” (high in many centralities relative to the best scorer) vs “specialized hubs” (high in only one)?
Solution: Compute multiple centralities, normalize them, and classify nodes by how many centralities place them in the top tier.
Query Code
Why It’s Interesting
Centrality is multifaceted — Degree ≠ betweenness ≠ closeness ≠ PageRank
Versatile hubs are robust — High across many metrics means genuine importance
Specialized hubs reveal roles — High in one metric reveals specific structural position
Example Output
Running on communication_network (email layer):
Node |
Degree |
Betweenness |
Closeness |
PageRank |
Versatility |
Type |
|---|---|---|---|---|---|---|
Manager |
9 |
1.0 |
1.0 |
0.4676 |
4 |
versatile_hub |
Dev1 |
1 |
0.0 |
0.5294 |
0.0592 |
0 |
peripheral |
Dev2 |
1 |
0.0 |
0.5294 |
0.0592 |
0 |
peripheral |
Interpretation: Manager is a versatile hub (it reaches at least 70% of the best score in all four centralities). All other nodes are peripheral in this star-topology email network.
DSL Concepts Demonstrated
.compute("degree", "betweenness_centrality", "closeness_centrality", "pagerank")— Compute multiple centralitiesNormalizing centralities for comparison
Derived metrics (versatility score)
Classification based on computed attributes
8. Edge Grouping and Coverage
Problem: You want to analyze which edges (connections) are important within and between layers. Which edges consistently appear in the top-k across different layer-pair contexts?
Solution: Use the new .per_layer_pair() method to group edges by (src_layer, dst_layer) pairs, then keep the top-k edges per pair. (Add .coverage(...) if you later need to filter across groups.)
Query Code
Why It’s Interesting
Layer-pair-aware analysis — Different layer pairs may have very different edge patterns
Universal edges — Edges important across multiple contexts are more robust
Cross-layer dynamics — Reveals how connections vary between intra-layer and inter-layer contexts
Edge-centric view — Complements node-centric analyses like hub detection
Example Output
Running on social_work_network with k=3 (insertion order per layer determines which edges are kept):
Edges Grouped by Layer Pair (top 3 per pair):
Source |
Target |
Source Layer |
Target Layer |
|---|---|---|---|
Alice |
Bob |
social |
social |
Alice |
Charlie |
social |
social |
Bob |
Charlie |
social |
social |
Alice |
Bob |
work |
work |
Alice |
David |
work |
work |
Alice |
Frank |
work |
work |
Alice |
Charlie |
family |
family |
Bob |
Eve |
family |
family |
David |
Frank |
family |
family |
Group Summary:
Source Layer |
Target Layer |
# Edges |
|---|---|---|
social |
social |
3 |
work |
work |
3 |
family |
family |
3 |
Interpretation: The query reveals edge distribution across layer pairs. Each intra-layer pair (e.g., social-social, work-work) contains up to k=3 edges. The sample dataset has only intra-layer edges; inter-layer pairs would appear if your network contains cross-layer connections. The family layer has sparser connectivity overall, so only three family edges remain after limiting.
When no sort key is provided, top_k keeps edges by their existing order; specify a weight to make the selection explicitly score-based.
DSL Concepts Demonstrated
.per_layer_pair()— Group edges by (src_layer, dst_layer) pairs.top_k(k, "weight")— Select top-k items per group.coverage(mode="at_least", k=2)— Optional cross-group filtering.group_summary()— Get aggregate statistics per groupEdge-specific grouping metadata in
QueryResult.meta["grouping"]
Tip
New in DSL v2
Edge grouping and coverage are new features that parallel the existing node
grouping capabilities. Use .per_layer_pair() for edges and .per_layer()
for nodes. Both support the same coverage modes and grouping operations.
9. Layer Algebra Filtering
Problem: You want to query specific subsets of layers using set operations. For instance, you might want to analyze “all layers except coupling layers” or “the union of biological layers.”
Solution: Use the LayerSet algebra with set operations (union, intersection, difference, complement) for expressive layer filtering.
Query Code
Why It’s Interesting
Expressive layer selection — Combine layers using set operations rather than listing them individually
Reusable layer groups — Define named layer groups for consistent reuse across queries
Exclude infrastructure layers — Easily filter out meta-layers like coupling layers
Complex filter expressions — Build sophisticated layer filters with union, intersection, difference
Example Output
The query returns a dictionary with multiple DataFrames showing different layer selection strategies:
No Coupling Layers: All layers except the coupling layer (useful for excluding meta-layers)
Bio Layers: Named group containing biological layers (ppi | gene | disease)
Intersection: Nodes appearing in both social AND work layers
Complement: Complement of coupling layer (equivalent to * - coupling)
DSL Concepts Demonstrated
L["* - coupling"]— Layer difference: all layers except couplingL["social & work"]— Layer intersection: nodes in both layersL["ppi | gene | disease"]— Layer union: combine multiple layers~LayerSet("coupling")— Layer complementL.define("bio", LayerSet(...))— Named layer groups for reuseString expression parsing for complex layer filters
10. Cross-Layer Paths with Algebra
Problem: When computing paths in multilayer networks, you may want to exclude certain layers (like coupling layers) that create artificial shortcuts, revealing more semantically meaningful paths.
Solution: Use layer algebra in path queries to control which layers participate in path computation.
Query Code
Why It’s Interesting
Avoid artificial shortcuts — Coupling layers often create paths that aren’t semantically meaningful
Compare path strategies — See how layer filtering affects connectivity
Layer-aware path finding — Control which layers participate in path computation
Semantic path discovery — Find paths that make sense in your domain
Example Output
The query returns a dictionary comparing path exploration with and without filtering:
All Layers: Node count and layer distribution when all layers are included
Filtered Layers: Node count and layer distribution excluding coupling layers
Interpretation: Excluding coupling layers often reveals more semantically meaningful paths by avoiding artificial shortcuts created by infrastructure layers.
DSL Concepts Demonstrated
Layer algebra in path queries
Comparing results with different layer subsets
Layer distribution analysis
Semantic path filtering
11. Null Model Comparison
Problem: How do you know if observed network patterns are statistically significant or just random? You need to compare actual network statistics against null model baselines.
Solution: Generate null models (e.g., configuration model) that preserve certain properties while randomizing connections, then compute z-scores to identify significant patterns.
Query Code
Why It’s Interesting
Statistical rigor — Establish baselines for significance testing
Identify exceptional patterns — Find nodes/structures that exceed random expectations
Configuration model — Preserves degree sequence but randomizes connections
Z-score analysis — Quantify how many standard deviations from expected
Essential for scientific conclusions — Avoid claiming significance for random patterns
Example Output
Running on a multilayer network returns a DataFrame with columns:
Node ID |
Layer |
Observed Degree |
Expected Degree |
Z-Score |
Is Significant |
|---|---|---|---|---|---|
Alice |
social |
5 |
3.2 |
2.8 |
True |
Bob |
social |
4 |
3.5 |
0.7 |
False |
Charlie |
work |
6 |
2.8 |
3.5 |
True |
Interpretation: Nodes with |z-score| > 2.0 are statistically significant (p < 0.05). Alice and Charlie have significantly higher degree than expected by chance, while Bob’s degree is within random variation.
DSL Concepts Demonstrated
Integration of null models with DSL queries
Statistical hypothesis testing
Computing z-scores and significance flags
Configuration model preserves degree distribution
Bootstrap resampling for confidence intervals
Note
Performance Note
This example uses 50 null model samples for CI speed. Production analyses typically use 100-1000 samples for more robust statistics.
12. Bootstrap Confidence Intervals
Problem: When analyzing centrality in multilayer networks, how stable are the measurements? Do nodes maintain consistent importance across layers, or is their centrality highly variable?
Solution: Analyze cross-layer variability to estimate uncertainty in centrality measures, identifying nodes with stable vs fragile importance.
Query Code
Why It’s Interesting
Quantify uncertainty — Know how reliable your centrality measurements are
Cross-layer variability — See which nodes maintain importance across contexts
Avoid over-interpretation — Don’t claim significant differences for small variations
Robust vs fragile patterns — Identify nodes with consistent vs inconsistent centrality
No distributional assumptions — Works when analytical standard errors unavailable
Example Output
Running on a multilayer network returns:
Node ID |
Layer |
Degree |
Mean Across Layers |
Std Dev |
Relative Variability |
Layer Coverage |
|---|---|---|---|---|---|---|
Alice |
social |
5 |
4.3 |
1.2 |
0.28 |
3 |
Bob |
work |
3 |
2.8 |
0.5 |
0.18 |
3 |
Charlie |
family |
2 |
3.1 |
1.8 |
0.58 |
2 |
Interpretation:
Low relative variability (Bob: 0.18) — Consistent importance across layers
High relative variability (Charlie: 0.58) — Importance varies dramatically by context
Layer coverage — Number of layers where the node appears
DSL Concepts Demonstrated
Cross-layer metric aggregation
Coefficient of variation for relative variability
Statistical comparison across layers
Uncertainty quantification in multilayer networks
13. Uncertainty-Aware Ranking
Problem: Traditional rankings order nodes by a single metric (e.g., max centrality). But what if a node has high centrality in one layer but low in others? How do you account for consistency vs peak performance?
Solution: Compare multiple ranking strategies—by maximum value, by mean across layers, and by consistency (low variability)—to make uncertainty-aware decisions.
Query Code
Why It’s Interesting
Beyond single-layer analysis — Consider multilayer context in rankings
Consistent vs peak performers — Identify nodes with stable vs specialized importance
Decision-making under uncertainty — Choose ranking strategy based on use case
Reveals ranking sensitivity — See how rankings change with different strategies
Practical implications — Different strategies matter for different applications
Example Output
Running on a multilayer network returns:
Node ID |
Layer |
Betweenness |
Mean |
Rel. Variability |
Rank by Max |
Rank by Mean |
Rank by Consistency |
Rank Change |
|---|---|---|---|---|---|---|---|---|
Alice |
work |
0.45 |
0.38 |
0.25 |
1 |
1 |
1 |
0 |
Charlie |
social |
0.42 |
0.28 |
0.52 |
2 |
3 |
5 |
3 |
Bob |
family |
0.38 |
0.35 |
0.18 |
3 |
2 |
2 |
1 |
Interpretation:
Alice — Top-ranked by all strategies (consistent high performer)
Charlie — Ranks highly by max (rank 2) but poorly by consistency (rank 5) due to high variability
Bob — More consistent than peak performer (rank 3 by max, rank 2 by consistency)
Rank change — Large values indicate sensitivity to ranking strategy
Use Cases:
Rank by max: When you need top performers in any context
Rank by mean: When you want overall consistent importance
Rank by consistency: When you need reliable performance across all contexts
DSL Concepts Demonstrated
Cross-layer variability analysis
Multiple ranking strategies
Consistency scoring
Sensitivity analysis for rankings
Practical decision-making with uncertainty
Choosing a Ranking Strategy
High-stakes decisions: Use consistency ranking to avoid nodes with variable performance
Exploratory analysis: Use max ranking to find peak performers
General purpose: Use mean ranking for balanced assessment
Large rank changes: Investigate why nodes rank differently across strategies
Using the Query Zoo
Getting Started
Install py3plex (if not already installed):
pip install py3plex
Run a single query:
from examples.dsl_query_zoo.datasets import create_social_work_network from examples.dsl_query_zoo.queries import query_basic_exploration net = create_social_work_network(seed=42) result = query_basic_exploration(net) print(result)
Run all queries:
cd examples/dsl_query_zoo python run_all.py
Run tests:
pytest tests/test_dsl_query_zoo.py -v
Adapting Queries to Your Data
All queries are designed to work with any multi_layer_network object. To adapt:
Replace the dataset:
from py3plex.core import multinet # Load your own network my_network = multinet.multi_layer_network() my_network.load_network("mydata.edgelist", input_type="edgelist_mx") # Run any query result = query_cross_layer_hubs(my_network, k=10)
Adjust parameters:
kinquery_cross_layer_hubs— Number of top nodes per layerLayer names in filters — Replace
L["social"]with your layer namesCentrality thresholds — Adjust percentile cutoffs as needed
Extend queries:
All query functions are in
examples/dsl_query_zoo/queries.py. Copy, modify, and experiment!
Datasets
Three multilayer networks are provided:
social_work_network
Layers: social, work, family
Nodes: 12 people
Structure: Overlapping social circles with different connectivity patterns per layer
communication_network
Layers: email, chat, phone
Nodes: 10 people (Manager, Dev team, Marketing, Support, HR)
Structure: Star topology in email, distributed in chat/phone
transport_network
Layers: bus, metro, walking
Nodes: 8 locations (CentralStation, ShoppingMall, Park, etc.)
Structure: Bus covers most locations, metro is faster but selective, walking is local
All datasets use fixed random seeds (seed=42) for reproducibility.
Further Reading
How to Query Multilayer Graphs with the SQL-like DSL — Complete DSL reference with syntax and operators
Multilayer Networks 101 — Theory of multilayer networks
DSL Reference — Full DSL grammar and API reference
Quick Start Tutorial — Quick start tutorial
Contributing Queries
Have an interesting multilayer query pattern? Contribute it to the Query Zoo!
Add your query function to
examples/dsl_query_zoo/queries.pyAdd tests to
tests/test_dsl_query_zoo.pyUpdate this documentation page
Submit a pull request!
See Contributing to py3plex for details.